Tutorial¶
Simple pendulum: build a surrogate model¶
In this tutorial we build a surrogate model for the damped simple pendulum using Arby. The physical setting is shown below.
This system can be described by the ordinary differential equation (ODE)
We can decompose the 2nd order ODE above in two 1st order ODEs as
Solving the pendulum equations¶
First we import some Python packages for handling and visualizing data.
[2]:
import numpy as np
[4]:
import matplotlib.pyplot as plt
Also we need an ODE solver. We use a module from the SciPy Python package.
[5]:
from scipy.integrate import odeint
Let’s define the ODE system and initial conditions
[6]:
def pend(y, t, b, λ):
θ, ω = y
dydt = [ω, -b*ω - λ*np.sin(θ)]
return dydt
[7]:
# set friction strength
b = 0.2
# set initial conditions
y0 = [np.pi/2, 0.]
[8]:
# set a time discretization
times = np.linspace(0,50,1001)
[9]:
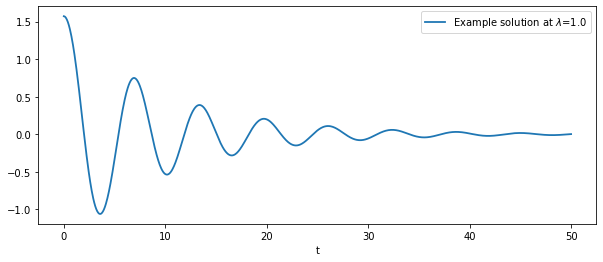
# plot a simple solution
λ = 1.
sol = odeint(pend,y0, times, (b,λ))
[10]:
plt.figure(figsize=(10,4))
plt.plot(times, sol[:,0], label=f'Example solution at $\lambda$={λ}', lw=1.8)
plt.xlabel('t')
plt.legend()
[10]:
<matplotlib.legend.Legend at 0x7f23dfcfad90>
Building the surrogate¶
Import the Arby main class for building surrogates
[11]:
from arby import ReducedOrderModel as ROM
and define a discretization of the parametric domain. We chose the domain for \(\lambda\) as \([1, 5]\).
[15]:
param = np.linspace(1,5,101)
The next step is to build a training set.
[16]:
training = []
for λ in param:
sol = odeint(pend,y0, times, (b,λ))
training.append(sol[:,0])
So far we have the main ingredients to buil a surrogate for the pendulum. For this, create a pendulum model as a ROM object
[17]:
pendulum = ROM(training, times, param, greedy_tol=1e-14, poly_deg=5)
and simply call/build the surrogate for some parameter value
[18]:
pendulum.surrogate(1.14)
[18]:
array([ 1.57079633, 1.56937605, 1.56513412, ..., -0.00282289,
-0.00331644, -0.00379569])
For the sake of brevity, name the surrogate.
[19]:
surr = pendulum.surrogate
We want to use the surrogate to make predictions. So lets look for an out-of-sample parameter to predict a solution
[20]:
param
[20]:
array([1. , 1.04, 1.08, 1.12, 1.16, 1.2 , 1.24, 1.28, 1.32, 1.36, 1.4 ,
1.44, 1.48, 1.52, 1.56, 1.6 , 1.64, 1.68, 1.72, 1.76, 1.8 , 1.84,
1.88, 1.92, 1.96, 2. , 2.04, 2.08, 2.12, 2.16, 2.2 , 2.24, 2.28,
2.32, 2.36, 2.4 , 2.44, 2.48, 2.52, 2.56, 2.6 , 2.64, 2.68, 2.72,
2.76, 2.8 , 2.84, 2.88, 2.92, 2.96, 3. , 3.04, 3.08, 3.12, 3.16,
3.2 , 3.24, 3.28, 3.32, 3.36, 3.4 , 3.44, 3.48, 3.52, 3.56, 3.6 ,
3.64, 3.68, 3.72, 3.76, 3.8 , 3.84, 3.88, 3.92, 3.96, 4. , 4.04,
4.08, 4.12, 4.16, 4.2 , 4.24, 4.28, 4.32, 4.36, 4.4 , 4.44, 4.48,
4.52, 4.56, 4.6 , 4.64, 4.68, 4.72, 4.76, 4.8 , 4.84, 4.88, 4.92,
4.96, 5. ])
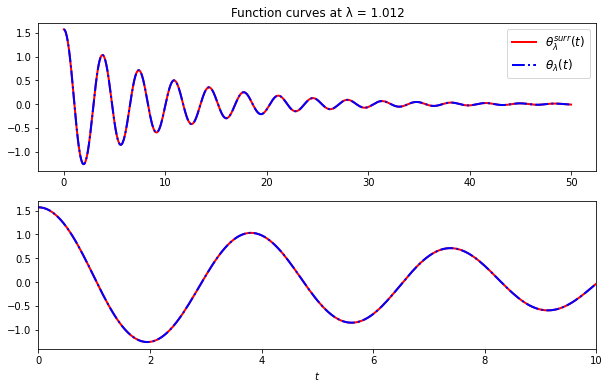
We choose par = 3.42. Now compare prediction and true solution.
[21]:
par = 3.42
sol = odeint(pend,y0, times, (b,par))[:,0]
fig, ax = plt.subplots(2,1, figsize=(10,6))
ax[0].plot(times, surr(par), 'r', lw=2, label='$θ_λ^{surr}(t)$')
ax[0].plot(times, sol, 'b-.', lw=2, label='$θ_λ(t)$')
ax[1].plot(times, surr(par), 'r', lw=2, label='$θ_λ^{surr}(t)$')
ax[1].plot(times, sol, 'b-.', lw=2, label='$θ_λ(t)$')
ax[1].set(xlim=(0,10))
ax[1].set(xlabel='$t$')
ax[0].set_title('Function curves at λ = 1.012')
ax[0].legend(fontsize = 'large')
[21]:
<matplotlib.legend.Legend at 0x7f23db2fe670>
Perform a simple profiling evaluating computation times for surrogate evaluations and for solving the ODE system at some parameter
[22]:
λ = 2.17
%timeit -t sol = odeint(pend,y0, times, (b,λ))
5.84 ms ± 389 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
[23]:
%timeit -t pendulum.surrogate(λ)
1.41 ms ± 95.5 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
We see that the surrogate is roughly 4 times faster than solving the ODE system
Benchmarks¶
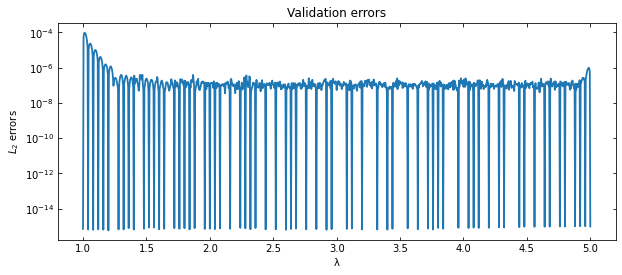
Perform a benchmark measuring the \(L_2\) relative error between the surrogate and a dense validation or test set of true solutions
where
[25]:
# name the norm tool from the integration object
norm = pendulum.basis_.integration.norm
# create a dense validation set (10x the size of the training set)
param_val = np.linspace(1,5,1001)
# compute errors
errors = []
for λ in param_val:
sol = odeint(pend,y0, times, (b,λ))[:,0]
errors.append(norm(sol - surr(λ))/norm(sol))
Plot errors
[26]:
fig, ax = plt.subplots(figsize=(10,4))
ax.plot(param_val, errors, lw=1.8)
ax.set_yscale('log')
ax.yaxis.set_ticks_position('both')
ax.tick_params(direction='in', which='both')
ax.xaxis.set_ticks_position('both')
ax.set_xlabel('λ')
ax.set_ylabel('$L_2$ errors')
ax.set_title('Validation errors')
[26]:
Text(0.5, 1.0, 'Validation errors')
Which is the parameter corresponding to the largest error?
[27]:
worst_lambda = param_val[np.argmax(errors)]
[28]:
# plot the overlap between the surrogate and the true solution model
# at the worst parameter
par = worst_lambda
sol = odeint(pend,y0, times, (b,par))[:,0]
fig, ax = plt.subplots(2,1, figsize=(10,6))
ax[0].plot(times, surr(par), 'r', lw=2, label='$θ_λ^{surr}(t)$')
ax[0].plot(times, sol, 'b-.', lw=2, label='$θ_λ(t)$')
ax[1].plot(times, surr(par), 'r', lw=2, label='$θ_λ^{surr}(t)$')
ax[1].plot(times, sol, 'b-.', lw=2, label='$θ_λ(t)$')
ax[1].set(xlim=(0,10))
ax[1].set(xlabel='$t$')
ax[0].set_title(f'Function curves at λ = {worst_lambda}')
ax[0].legend(fontsize = 'large')
[28]:
<matplotlib.legend.Legend at 0x7f23d9cbb4c0>
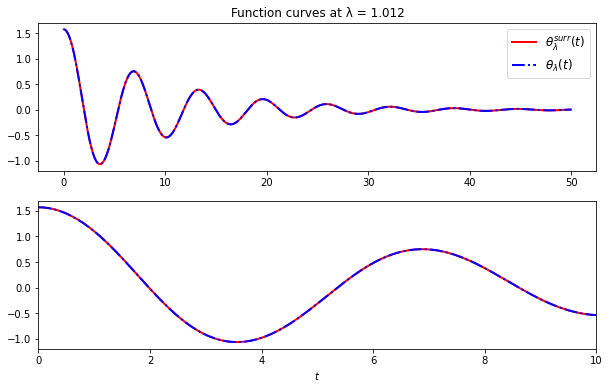
We see from this plot that the curves are indistinguishable to eyeball resolution
Build a reduced basis¶
Lets go deeper. The Reduced Basis Method is a reduced order modeling technique for building a near-optimal basis of functions that spans the training set at an user-specified tolerance. The basis is built by iteratively choosing those training functions which best represent the entire set. In this way, as opposed to other dimensional reduction techniques such as Proper Orthogonal Decomposition, the reduced basis is directly interpretable since it is built out from training functions. Another kindness of this approach is that whenever we want more accuracy we just add more basis elements to the computed one: the construction is hierarchical.
Suppose we have a training set \(\{f_{\lambda_i}\}_{i=1}^N\) of parameterized real functions. This set may represent a non-linear model, perhaps solutions to PDEs. We would like, if possible, to reduce the dimensionality/complexity of it by finding a compact representation in terms of linear combinations of basis elements \(\{e_i\}_{i=1}^n\), that is,
f is an arbitrary training function and the \(c_i\)’s are the projection coefficients \(<e_i,f>\) computed in some inner product \(<\cdot,\cdot>\) on the space of functions. The RB method chooses a set of optimal functions belonging to the training set itself which defines a finite dimensional subspace capable to represent the entire training set up to a user-specified tolerance.
To build a reduced basis with Arby, you just provide the training set of functions and the discretization of the physical variable \(x\) to the reduced_basis function. The later is to define the integration scheme that is used for computing inner products. For the Bessel example,
[ ]:
from arby import reduced_basis
rb_data = reduced_basis(training_set=training,
physical_points=times, greedy_tol=1e-12)
The greedy_tol parameter is the accuracy in the \(L_2\)-norm that our reduced basis is expected to achieve. The output rb_data contains all the relevant information related to greedy calculations. It contains a basis object which comprises the reduced basis and several utilities for interacting with it. The other outputs are the greedy errors and indices, and the projection_matrix, which stores projection coefficients built in the greedy algorithm. For example, to
call the reduced basis array do
[ ]:
rb_data.basis.data
The reduced basis is an orthonormalized version of the set of functions selected by the greedy algorithm from training data, which are indexed by the greedy indices. You can obtain those functions by filtering the training set
[ ]:
training_set[rb_data.indices]
For conditioning purposes, the greedy algorithm orthonormalizes these functions using the inner product implied by the integration scheme.
The number of basis elements rb_data.basis.Nbasis_ represents the dimension of the reduced space. It is not a fixed quantity since we change it by modifying the greedy tolerance. The lower the tolerance, the bigger the number of basis elements needed to reach that accuracy. With Arby, we can tune the accuracy of the reduced basis through the greedy_tol parameter.
To measure the effectiveness of the reduced basis in approximating the training set just compute the norm of difference between a training function f and its projected version using the tools coming inside the rb_data.basis class object.
[ ]:
projected_f = rb_data.basis.project(f)
norm = rb_data.basis.integration.norm
L2_error = norm(f - projected_f)
Or take a shortcut by doing
[ ]:
projection_error = rb_data.basis.projection_error
squared_L2_error = projection_error(f)
The output is the square version of the error computed in the previous code block.
Further reading¶
This package was built upon previous work made in the field of Gravitational Wave (GW) science. Important papers are
Towards beating the curse of dimensionality for gravitational waves using Reduced Basis
Fast prediction and evaluation of gravitational waveforms using surrogate models
For some theoretical aspects on the Reduced Basis greedy algorithm and the Empirical Interpolation Method check
For a thorough review on Reduced Order Methods as implemented in this package, check
For awsome visuals on the application of surrogate modeling in GW Science check this.